|
|
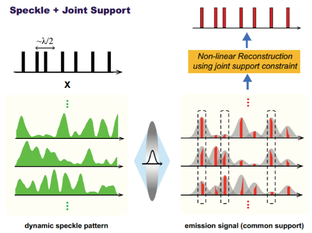
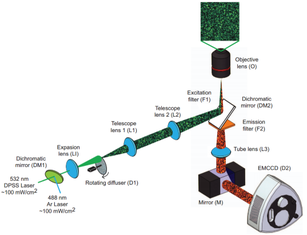
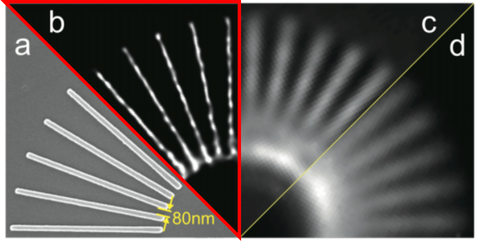
Nanoscopy using speckle illumination and joint support recoveryJ. Min, J.D. Jang, D.M. Keum, S.W. Ryu, C.H. Choi, K.H. Jeong, and J.C. Ye "Fluorescent microscopy beyond diffraction limits using speckle illumination and joint support recovery", Scientific Reports, June, 2013
A primary limitation of fluorescence microscopy is that it cannot resolve sub-cellular structures beyond the diffraction limit. In many biological experiments, this resolution limit is not satisfactory for investigating the structures and functions of macromolecules. In order to address this problem, super-resolution far-field fluorescence microscopy has been investigated extensively for the past decade. A foundational concept behind this super-resolution microscopy is to exploit non-linear optical phenomena. For example, in stimulated emission depletion (STED) microscopy, a Gaussian-shaped excitation beam and a red-shifted doughnut-shaped STED beam are used to sharpen the effective point spread function (PSF) by quenching excited fluorophores at the rim of the PSF. Unlike STED, stochastic optical reconstruction microscopy (STORM) and photoactivated localization microscopy (PALM) are based on the single molecule imaging principle by localizing the sparsely activated fluorophores using photoswitchable probes. However, considering that the large inventories of the existing fluorophores or fluorescent proteins are not photoswitchable, a new super-resolution imaging architecture that is compatible with the conventional experimental protocols and fluorescence probes should be developed in order to increase the use of super-resolution microscopy in biological experiments. We proposed a novel speckle illumination microscopy technique that overcomes the diffraction limit by exploiting the minimal requirement that is common for all the existing super-resolution microscopy, i.e. that the fluorophores’ locations do not vary during the acquisition time. Using numerical and real experiments, we demonstrate that the proposed method can improve the resolution up to threefold. Significance The proposed method succeeds for standard fluorescence probes and experimental protocols, it can be applied in routine biological experiments. Moreover, it can be easily implemented by modifying a convention fluorescence microscopy without a sophisticated optical alignment. The spatial resolution of the proposed method is not limited by a factor of 2, whereas structure illumination microscopy (SIM) only improves the optical resolution two times by synthesizing an extended aperture from multiple images with distinct spatial modulations. Fast localization algorithm for high-density super-resolution microscopy data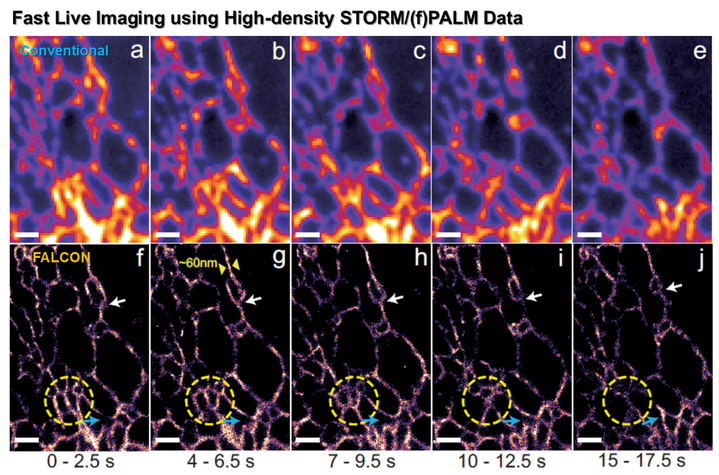 FALCON performance on live ER data. Live imaging of the endoplasmic reticulum protein, reticulon-4 fused to tdEos imaged over 20 s in a U2OS cell. FALCON performance on live ER data. Live imaging of the endoplasmic reticulum protein, reticulon-4 fused to tdEos imaged over 20 s in a U2OS cell.
J. Min, C. Vonesch, H.Kirshner, L. Carlini, N. Olivier, S. Holden, S. Manley, J.C. Ye, M. Unser, " FALCON: fast and unbiased reconstruction of high-density super-resolution microscopy data," Scientific Reports , Apr. 2014.
Single-molecule localization microscopy methods, such as STORM and (F)PALM utilize sparse activation of photo-switchable fluorescent probes in both temporal and spatial domain. Each activated probe can be assimilated to an ideal point source so that the acquired images consist of isolated replicates of the point spread function of the microscope (PSF). This allows one to achieve sub-pixel accuracy on the order of tens of nanometers for the estimated location of each probe. In general, reconstruction of sub-cellular structures relies on numerous localized probes, and the required acquisition time of these methods is therefore relatively long, i.e. on the order of minutes. This is a serious limitation when investigating live-cell dynamics. We propose an algorithm for high-density super-resolution microscopy which combines a sparsity-promoting formulation with Taylor series approximation of the PSF. Our algorithm is designed to provide unbiased localization on continuous space and high recall rates for high density imaging, and to have orders-of-magnitude faster run time compared to previous high-density algorithms. We validated our algorithm on both simulated and experimental data, and demonstrated live-cell imaging with temporal resolution of 2.5 s. by recovering fast ER dynamics. Significance All previous high-density localization algorithm using sparsity-promoting priors are based on similar discrete formulations. Such formulations, however, have inherent limitations. First, discrete-domain formulations can account for only pre-defined locations, not all possible probe locations over a continuum. Second, using a finer sub-pixel grid increases the computational load, especially with the linear-programming approach used in CSSTORM. To address these limitations of the current high-density localization algorithms, we proposed a FAst Localization algorithm based on a CONtinuous-space formulation (FALCON) for high density super-resolution microscopy data. In particular, to obtain a grid-free reconstruction, our approach combines a sparsity-promoting formulation with a Taylor approximation of the PSF. In addition, implemented in a computationally efficient way by utilizing a fast compressed sensing algorithm called ADMM (Alternating Direction Method of Multipliers). |
|
ABOUT US
Our research activities are primarily focused on the signal processing and machine learning for high-resolution high-sensitivity image reconstruction from real world bio-medical imaging systems. Such problems pose interesting challenges that often lead to investigations of fundamental problems in various branches of physics, mathematics, signal processing, biology, and medicine. While most of the biomedical imaging researchers are interested in addressing this problem using off-the-self tools from signal processing, machine learning, statistics, and optimization and combining their domain-specific knowledge, our approaches are unique in the sense that I believe that actual bio-medical imaging applications are a source of endless inspiration for new mathematical theories and we are eager to solve both specific applications and application-inspired fundamental theoretical problems.
|
CONTACT US
Bio Imaging. Signal Processing & Learning
Graduate School of AI KAIST 291 Daehak-ro, Yuseong-gu Daejeon 305-701, Korea Copyright (c) 2014, BISPL All Rights Reserved. |
|